
Developed by Tashrif Billah, Sylvain Bouix, and Yogesh Rathi, Brigham and Women’s Hospital (Harvard Medical School).
This pipeline is also available as Docker and Singularity containers. See pnlpipe-containers for details.
Table of Contents
- Citation
- Introduction
- Dependencies
- Installation
- Multiprocessing
- Pipeline scripts overview
- Tutorial
- Support
Citation
If this pipeline is useful in your research, please cite as below:
Billah, Tashrif; Bouix, Sylvain; Rathi, Yogesh; NIFTI MRI processing pipeline, https://github.com/pnlbwh/pnlNipype, 2019, DOI: 10.5281/zenodo.3258854
Introduction
pnlNipype is a Python-based framework for processing anatomical (T1, T2) and diffusion weighted images. It comprises some of the PNL’s neuroimaging pipelines. A pipeline is a directed acyclic graph (DAG) of dependencies. The following diagram depicts functionality of the NIFTI pipeline, where each node represents an output, and the arrows represent dependencies:
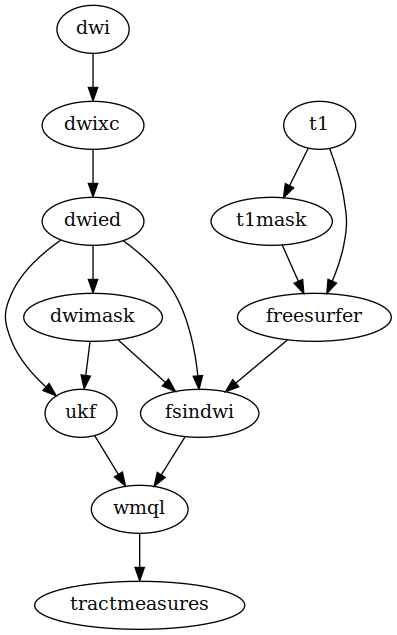
Detailed DAGs are available here.
Dependencies
- dcm2niix
- ANTs >= 2.3.0
- UKFTractography >= 1.0
- FreeSurfer >= 5.3.0
- FSL >= 5.0.11
- Python >= 3.6
Installation
1. Install prerequisites
Installing pnlNipype requires you to install each of the dependencies independently. This way, you can have more control over the requisite software modules. The independent installation is for users with intermediate programming knowledge.
Install the following software (ignore the one(s) you have already):
- Python >= 3.6
- FreeSurfer >= 5.3.0
- FSL >= 5.0.11
Check system architecture
uname -a # check if 32 or 64 bit
Python 3
Download Miniconda Python 3.6 bash installer (32/64-bit based on your environment):
sh Miniconda3-latest-Linux-x86_64.sh -b # -b flag is for license agreement
Activate the conda environment:
source ~/miniconda3/bin/activate # should introduce '(base)' in front of each line
FSL
Follow the instruction to download and install FSL.
FreeSurfer
Follow the instruction to download and install FreeSurfer >= 5.3.0
After installation, you can check FreeSurfer version by typing freesurfer on the terminal.
Conda environment
git clone --recurse-submodules https://github.com/pnlbwh/pnlNipype.git && cd pnlNipype
# install the python packages required to run pnlNipype
conda create -y -n pnlpipe9 -c conda-forge --override-channels python
conda activate pnlpipe9
pip install -r requirements.txt
T1 and T2 training masks
(Optional) In the past, we used MABS (Multi Atlas Brain Segmentation) method to generate T1 and T2 masks. But now we use HD-BET to generate those. Hence, this step remains optional. But if you want, you can ‘install’ our training masks:
# define PYTHONPATH so that pnlNipype/cmd/install.py can find pnlpipe_software/* installation scripts
export PYTHONPATH=`pwd`
# define PNLPIPE_SOFT as the destination of training masks
export PNLPIPE_SOFT=/path/to/wherever/you/want/
# training data for MABS mask
cmd/install.py trainingDataT1AHCC
cmd/install.py trainingDataT2Masks
unset PYTHONPATH
However, other external software should be built from respective sources:
- ANTs
You can build ANTs from source. Additionally, you should define ANTSPATH.
The other alternative can be installing ANTs through conda. We have developed a conda package for ANTs==2.3.0.
You can get that by:
conda install -c pnbwh ants
- dcm2niix
dcm2niix executable will create NIFTI file from DICOM. The pipeline uses a reliable converter dcm2niix. Building of dcm2niix is very straightforward and reliable. Follow this instruction to build dcm2niix.
- UKFTractography
Follow this instruction to download and install UKFTractography.
- tract_querier & whitematteranalysis
We found that tract_querier and whitematteranalysis dependencies do not quite agree with simpler pnlNipype dependecies. Hence, you may want to install them within a separate Python environment outside of pnlpipe3. Individual repository instructions can be followed to install them. It will be something in line of:
git clone http://github.com/demianw/tract_querier.git
cd tract_querier
pip install .
Special care has to be taken so that tract_querier’s and whitematteranalysis’s executables are
in PATH environment variable.
2. Configure your environment
It is hard to definitively suggest how to configure your environment. But it should be something akin to:
source ~/miniconda3/bin/activate # should introduce '(base)' in front of each line
export FSLDIR=/path/to/fsl/ # setup fsl environment
source $FSLDIR/etc/fslconf/fsl.sh
export PATH=$PATH:$FSLDIR/bin
export FREESURFER_HOME=/path/to/freesurfer # you may specify another directory where FreeSurfer is installed
source $FREESURFER_HOME/SetUpFreeSurfer.sh
# source external software
ANTSDIR=/path/to/ANTs
ANTSPATH=$ANTSDIR/build/ANTS-build/Examples
PATH=$ANTSPATH:$ANTSDIR/Scripts:$PATH # define ANTSPATH and export ANTs scripts in your path
PATH=/path/to/dcm2niix/build/bin:$PATH
PATH=/path/to/ukftractography/build/UKFTractography-build/UKFTractography/bin:$PATH
PATH=/path/to/pnlNipype/exec:$PATH
export ANTSPATH PATH
(If you would like, you may include the above in your bashrc to have environment automatically setup
every time you open a new terminal)
3. Temporary directory
Both pnlpipe and pnlNipype have centralized control over various temporary directories created down the pipeline.
The temporary directories can be large, and may possibly clog the default /tmp/ directory. You may define custom
temporary directory with environment variable PNLPIPE_TMPDIR:
mkdir /path/to/tmp/
export PNLPIPE_TMPDIR=/path/to/tmp/
4. Tests
Upon successful installation, you should be able to see help message of each script in the pipeline:
cd lib
scripts/atlas.py --help
scripts/fs2dwi.py --help
...
Multiprocessing
Multi-processing is an advanced feature of pnlNipype. The following scripts are able to utilize python multiprocessing capability:
atlas.py
pnl_eddy.py
pnl_epi.py
wmql.py
antsApplyTransformsDWI.py
You may specify N_PROC parameter in scripts/util.py for default number of processes to be used across scripts in the pipeline.
N_PROC = '4'
On a Linux machine, you should find the number of processors by the command lscpu:
On-line CPU(s) list: 0-55
You can specify any number not greater than the On-line CPU(s). However, one caveat is, other applications in your computer
may become sluggish or you may run into memory error due to heavier computation in the background. If this is the case,
reduce NCPU (--nproc) to less than 4.
Pipeline scripts overview
scripts is a directory of PNL specific scripts that implement various
pipeline steps. These scripts are the successors to the ones in pnlpipe
used for NRRD format data. Besides being more robust and up to date with respect to software such
as ANTS, they are implemented in python using
the shell scripting library plumbum.
Being written in python means they are easier to understand and modify,
and plumbum allows them to be
almost as concise as a regular shell script.
You can call any of these scripts directly, e.g.
scripts/align.py -h
It’s important to note that usually the scripts are calling other binaries, such as those in ANTS, FreeSurfer and FSL. So, make sure you source each of their environments so individual scripts are able to find them.
This table summarizes the scripts in pnlNipype/scripts/:
| Category | Script | Function |
|---|---|---|
| General | align.py | axis aligns and centers an image |
| General | bet_mask.py | masks a 3D/4D MRI using FSL bet |
| General | masking.py | skullstrips by applying a labelmap mask |
| General | maskfilter.py | performs morphological operation on a brain mask |
| General | resample.py | resamples a 3D/4D image |
| DWI | unring.py | Gibbs unringing |
| DWI | antsApplyTransformsDWI.py | applies a transform to a DWI |
| DWI | bse.py | extracts a baseline b0 image |
| DWI | pnl_epi.py | corrects EPI distortion via registration |
| DWI | fsl_topup_epi_eddy.py | corrects EPI distortion using FSL topup and eddy_openmp |
| DWI | pnl_eddy.py | corrects eddy distortion via registration |
| DWI | fsl_eddy.py | corrects eddy distortion using FSL eddy_openmp |
| DWI | ukf.py | convenient script for running UKFTractography |
| Structural | atlas.py | computes a brain mask from training data |
| Structural | makeAlignedMask.py | transforms a labelmap to align with another structural image |
| Structural | fs.py | convenient script for running freesurfer |
| Freesurfer to DWI | fs2dwi.py | registers a freesurfer segmentation to a DWI |
| Tractography | wmql.py | simple wrapper for tract_querier |
| Tractography | wmqlqc.py | makes html page of rendered wmql tracts |
The above executables are available as soft links in pnlNipype/exec directory as well:
| Soft link | Target script |
|---|---|
| fsl_eddy | ../scripts/fsl_eddy.py |
| fsl_toup_epi_eddy | ../scripts/fsl_topup_epi_eddy.py |
| masking | ../scripts/masking.py |
| nifti_align | ../scripts/align.py |
| unring | ../scripts/unring.py |
| maskfilter | ../scripts/maskfilter.py |
| resample | ../scripts/resample.py |
| nifti_atlas | ../scripts/atlas.py |
| nifti_bet_mask | ../scripts/bet_mask.py |
| nifti_bse | ../scripts/bse.py |
| nifti_fs | ../scripts/fs.py |
| nifti_fs2dwi | ../scripts/fs2dwi.py |
| nifti_makeAlignedMask | ../scripts/makeAlignedMask.py |
| nifti_wmql | ../scripts/wmql.py |
| pnl_eddy | ../scripts/pnl_eddy.py |
| pnl_epi | ../scripts/pnl_epi.py |
| ukf | ../scripts/ukf.py |
For example, to execute axis alignment script, you can do either of the following:
pnlNipype/exec/nifti_align -h
pnlNipype/scripts/align.py -h
Tutorial
See the TUTORIAL for workflow and function of each script.
Support
Create an issue at https://github.com/pnlbwh/pnlNipype/issues . We shall get back to you as early as possible.

